Methods
smFISH
Image Processing and Analysis
Nervous system dissections

Single-molecule Fluorescent In Situ Hybridisation is a method we use to label RNA in cells and intact tissues.
Nervous system dissections
Image Processing and Analysis
Nervous system dissections

We use the fly larval brain and neuromuscular junction as a model to study the nervous system. Dissecting these small tissues can be challenging
Image Processing and Analysis
Image Processing and Analysis
Image Processing and Analysis

Our expertise in imaging doesn't stop with acquisition, we have also developed and use methods of post-acquisition processing to improve our data quality.
RNA Localisation
Image Processing and Analysis

Where RNA is in cells and how that impacts on development and nervous system plasticity and function is a key interest in the Davis Lab
Fly Embryos and Oocytes
Fly Embryos and Oocytes

Before the Davis lab worked on the nervous system we studied development in the fly embryo and oocyte.
Downloads
Fly Embryos and Oocytes

Protocols and Methods papers from our lab for you to download

Single-molecule Fluorescent In Situ Hybridisation
Single-molecule RNA detection in cells and tissues
In the Davis lab we work not only in cell culture systems, but also in complex intact tissues of the fly nervous system. Keeping these tissues intact allows us to study these systems in a more realistic and true-to-life manner but doing so offers new challenges. As much of our work is concerned with the location of RNA, down to the single molecule level, locating single transcripts deep within tissues presents a series of challenges. These challenges occur not just in the difficulty of labelling RNA transcripts but also in imaging them. We always aim to improve and develop our smFISH and related imaging methods to ensure the highest quality data whether it's in a single cell layer or one hundred microns deep in a fly brain. For further details see our publications of optimised smFISH at the fly neuromuscular junction here and in the intact fly brain here
Nervous System Dissections

Brains and neuromuscular junctions
In the Davis lab, we primarily work on two tissues in the nervous system; The brain and the neuromuscular junction (NMJ). The brain offers us a complex tissue, analogous to the mammalian brain, with thousands of neurons and glia formed into discrete regions with different functions. The NMJ offers a highly tractable synaptic system that is open to chemical and electrical stimulation while also offering a wonderful model for synaptic plasticity that is very accessible for imaging.
Dissection
Dissecting out tissues like these from a small animal only a few millimetres long can be challenging, but there are established protocols to follow that can make this achievable after a relatively short period of instruction and practice.
Larval Brain Dissection
NMJ Dissection
Image Processing and Analysis
Taking Microscopy further
In the Davis lab we use the latest imaging technology to obtain microscopy data of the highest quality. In order to do this it's vital to be able to process and interpret these images after they've been acquired. We use a wide range of image processing and analysis tools, including many that we develop ourselves to answer the questions we are interested in.
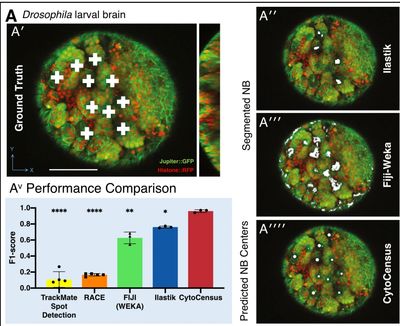
CytoCensus
Modern imaging datasets can be extremely complex and huge in scale, often to the point where analysis by a human would be difficult and take an extremely long time. We developed CytoCensus, a machine-learning based tool that is able to process large 4D datasets with minimal input from the user. CytoCensus is capable of detecting and counting particular cell types throughout the dataset in a fraction of the time it would take a human user, while outperforming other commercially available image analysis software.
Particle Stats
The study of dynamic cellular processes in living cells is central to biology and is particularly powerful when the motility characteristics of individual objects within cells can be determined and analysed statistically. However, commercial programs only offer a very limited range of inflexible analysis modules. We developed an open-source software to perform these functions and more.
RNA Localisation
More than just a messenger
RNA is well known for being the messenger molecule that bridges the gap between transcription and translation, but in the Davis lab we are very interested in what happens in between these two processes. We have worked extensively with labelled RNA to discover where it goes and how this impacts on development and synaptic plasticity. In order to do this we have developed and adapted methods to image RNA in both live and fixed material. The Davis lab were pioneers in detecting low levels of RNA in situ such as in this study where we investigated the localisation of transcribed genes in Drosophila nuclei.
Fly Embryos and Oocytes
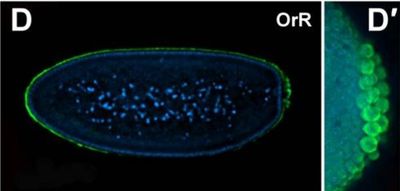
A wonderful model to study
Before we began studying the nervous system, the Davis Lab made great strides in understanding RNAs role in development by studying the embryos and oocytes of flies. This system is highly informative of how RNA plays a role in patterning and early development, while being relatively amenable to genetic manipulation and very accessible to modern imaging techniques.
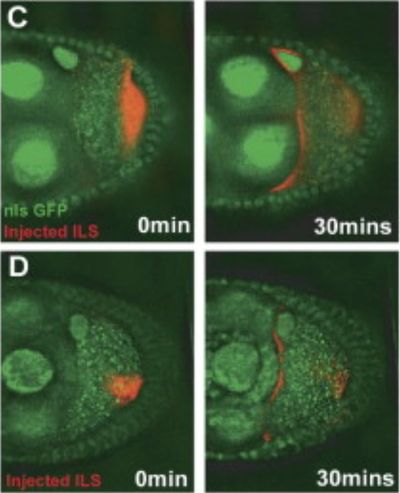
RNA Injection
A valuable advantage of the embryo and oocyte system is that injections of labelled RNA is possible without causing long term damage to the cells. This method requires some fine skills, but in the Davis lab we developed several methods that allowed us to use it to great effect in answering our questions.
Methods Papers and Protocols
Ribo-Pop: Simple, cost-effective ribosomal RNA depletion (pdf)
DownloadCytoCensus, mapping cell identity and division in tissues and organs using machine learning (pdf)
DownloadsmFISH at the neuromuscular junction (pdf)
DownloadLive Cell Imaging in Drosophila (pdf)
DownloadsmFISH in the fly larval brain (pdf)
DownloadDrosophila Macrophage Preparation and Screening (pdf)
DownloadMaking the message clear: visualizing mRNA localization. (pdf)
DownloadCollection and mounting of Drosophila embryos for imaging (pdf)
DownloadEarly sensitive FISH method (pdf)
DownloadHCRv3_probe_design_zenodo_manual: https://zenodo.org/records/12518278 (pdf)
Download
